 |
| Why GFP and DsRed2 cannot be
separated and What to use instead. |
To mark the expression of two different genes, each indepently and in vivo, GFP and DsRed2 may not be good choices.
Instead, consider using ECFP and DsRed2.
Why?
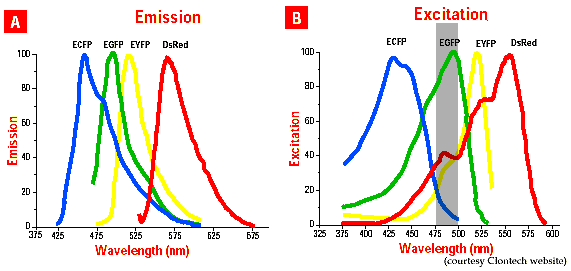
At first glance, the emission spectra of GFP and DsRed2 appear to have very little overlap which leads us to believe that this combination would be excellent for separating each signal (A).
However, the absorbtion specta for GFP and DsRed2 overlap significantly (gray zone in B). DsRed2 is visible through even the most narrow of the narrow band green emission filters.
For separation, use ECFP and DsRed2 instead. Although some DsRed2 is excited at 435 nm, which is the standard excitation for ECFP, according to the published spectra, no DsRed2 should be visible through the narrow ECFP emission filter. Also, we have found that ECFP absorbs well at 414 nm, which is outside the DsRed2 absorbtion range, to further assure uniqueness of signal.
(This section last revised 17 Sept 2001)
The vector encodes dsRed2 and the Clontech product is pdsRed2-N1.
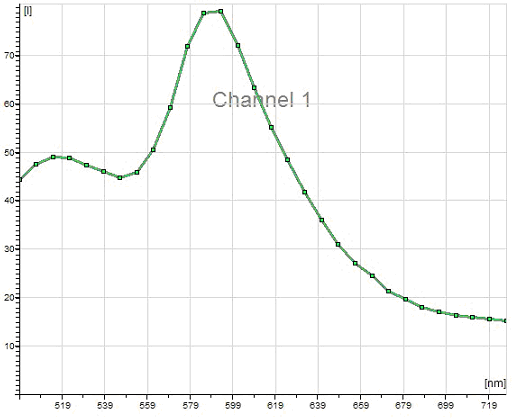
Excitation at 458 and 476 nm and spectra recorded with Leica
AOBS confocal on 6 Oct 2005.
(This section last revised 6 Oct 2005)